Development of an AlphaLisa assay for Screening Histone Methyltransferase Inhibitors (NB-SC107)
A top pharmaceutical company based in US approached us to develop an AlphaLisa assay for the histone methyltransferase G9a/ EHMT2. The experimental design and results are as follows:
Assay Buffer:
Used for the enzyme reaction and diluting G9a enzyme, biotinylated histone H3 (1-21) peptide, SAM and inhibitors.
Formulation: 50 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM DTT, 0.01% Tween-20, 0.005% BSA.
HMT Buffer 1:
Used to dilute the acceptor and donor beads.
Assay Protocol:
Dilute G9a enzyme, biotinylated histone H3 (1-21) peptide, SAM and inhibitors in assay buffer just before use.
Add the following to 384-well white plate wells:
5 μL assay buffer or inhibitor (2X)
2.5 μL of enzyme (4X)
2.5 μL of biotinylated Histone H3 (1-21) peptide/ SAM mix (4X)
Cover the plate with a sealer and incubate at RT.
Prepare H3K9me2 acceptor beads in 1X Epigenetics Buffer 1. Add 5 μL Acceptor beads to each well. Cover the plate with a sealer and incubate for 60 min at RT in dark.
Prepare streptavidin donor beads in 1X Epigenetics Buffer 1. Add 10 μL Donor beads in subdued light. Cover the plate with a sealer and incubate for 30 min at RT in dark.
Read signal in Alpha enabled readers. Novatein Biosciences used BMG Pherastar.
Effect of time on G9a enzyme activity:
G9a was incubated with 100 nM biotinylated Histone-H3 (1-21) peptide and 100 µM S-adenosyl methionine (SAM) for various time points. Acceptor and donor beads were used at 5 μg/ml and 20 μg/ml. As the counts obtained with 5 μg/ml were good, we used both the beads at 5 μg/ml final concentration in the assay.
|
Time (min) |
Alpha Counts (blank subtracted) |
||
|
180 |
2755792 |
2870244 |
2572242 |
|
120 |
2630541 |
2718970 |
2885052 |
|
60 |
1685737 |
1353978 |
1692881 |
|
30 |
905154 |
854907 |
967214 |
|
15 |
172824 |
80978 |
204934 |
|
5 |
14535 |
6422 |
17062 |
|
0 |
0 |
0 |
0 |
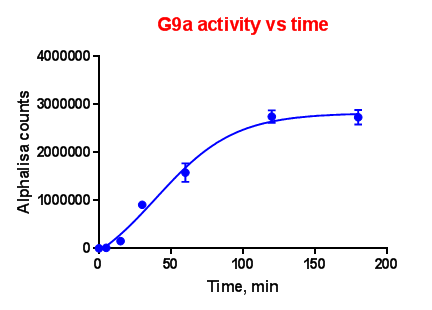
For all the subsequent experiments, enzyme assay incubation was carried out for 1 hour.
Effect of G9a concentration on enzyme activity:
G9a was incubated with biotinylated Histone-H3 (1-21) peptide and S-adenosyl methionine (SAM) for 1 hour. Acceptor and donor beads were used at 5 μg/ml. Beads alone was used as blank.
|
G9a (nM) |
Alpha counts (blank subtracted) |
||
|
0.05 |
5985 |
8417 |
3401 |
|
0.1 |
83733 |
98553 |
129029 |
|
0.5 |
2049663 |
1785278 |
2162409 |
|
1 |
2999625 |
3370296 |
3142182 |
|
3.6 |
4724996 |
4659807 |
4927650 |
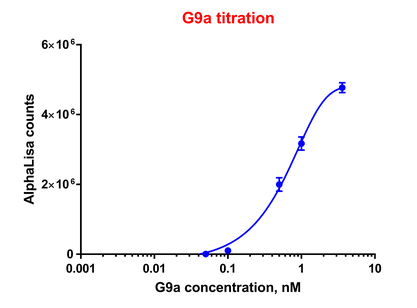
For all the subsequent experiments, 0.5 nM G9a was used.
Effect of SAM concentration on enzyme activity:
G9a was incubated with biotinylated Histone-H3 (1-21) peptide and S-adenosyl methionine (SAM) for 1 hour. Acceptor and donor beads were used at 5 μg/ml.
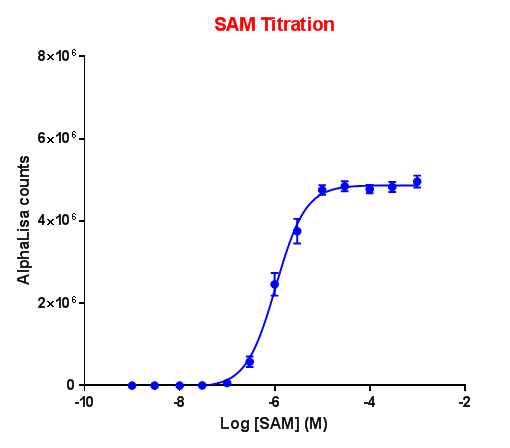
For all the subsequent experiments, ~Km concentration of SAM was used.
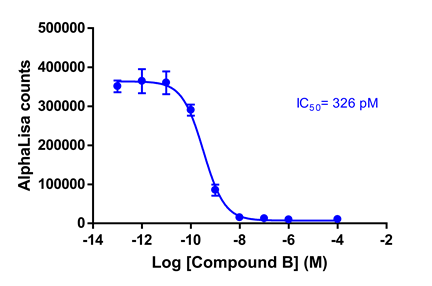
Inhibitor titration:
Two compounds were given to us to evaluate in this assay setting:
Compounds (100 pM – 100 µM) were pre-incubated with G9a for 15 min at RT. Biotinylated Histone H3 (1-21) peptide and SAM were added and further incubated for 1 hour at RT. Acceptor and donor beads were added at 5 μg/ml and read on BMG Pherastar. IC50 was calculated using Graphpad Prism.
|
Compound A (M) |
AlphaLisa counts |
||
|
0 |
264765 |
244865 |
273171 |
|
1e-010 |
391874 |
382441 |
356612 |
|
1e-009 |
363907 |
330334 |
335046 |
|
1e-008 |
283283 |
292220 |
326192 |
|
1e-007 |
179702 |
140330 |
165547 |
|
0.000001 |
53143 |
37221 |
44270 |
|
0.00001 |
14535 |
12103 |
12198 |
|
0.0001 |
8987 |
7714 |
8645 |
IC50 of compound A was determined as 7.1 nM.